Thanks @fxbuson to create this pTI expression plasmid topic 
Also, is good to know you have shared it to other labs and would be awesome to have some feedback from them.
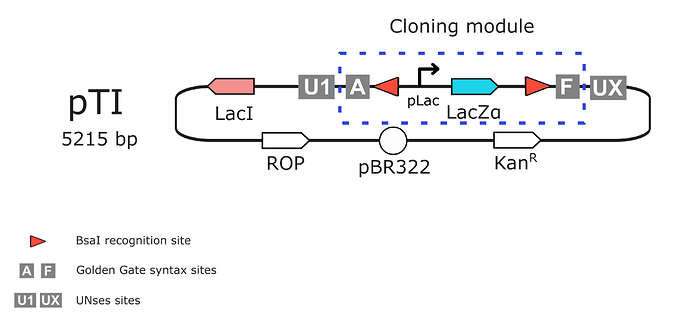
pTI general information
(plasmid sequence available here)
As mentioned before this plasmid was designed as a golden gate BsaI acceptor and expression vector. It includes:
i) Cloning module: composed by a LacZ expression cassette which is removed after an effective golden gate assembly  in a blue/white assay, colonias which doesn’t insert the assembled enzyme gene will be blue
in a blue/white assay, colonias which doesn’t insert the assembled enzyme gene will be blue  . It is flanked by BsaI recognition sites and A(GGAG) - F(CGCT) standard level 1 assembly syntax sequences.
. It is flanked by BsaI recognition sites and A(GGAG) - F(CGCT) standard level 1 assembly syntax sequences.
ii) Verification motifs: The clonning module is flanked by UNSes sequences (Torella et al., 2014) to allow standard sequencing verification (though U1F/UXR primers) or any other PCR downstream procedure of interest.
U1F: cattactcgcatccattctcaggctg
UXR: GGTGGAAGGGCTCGGAGTTGTGG
iii) LacI regulation: It has constitutive expression of LacI repressor to allow lac box regulated promoters (e.g. pLac or pT7_lac0) protein production, and enhance the T7RNAP regulation included in λDE3 lysogen (minimizing leacky expression of T7RNAP). The LacI gene expression is preciselly leveled to perform this tasks.
iv) Mainteinance elements: Kanamycin resistance cassette to perform the selection and keep the plasmid. Kanamycin usage agrees with the assembly plasmid hierachy requirements, is cheap, broadly used/available and has an optimal performance. Replication machinery is composed of pBR322 plus ROP gene to keep a low plasmid copy number.
v) Upper uLoop level assembly elements: Additionally, between cloning module and UNSes, there are uLoop even assembly schemas that could be used to include the enzyme expression cassette in downstream SapI multiple gene assemblies.
Mode of use
Once use this plasmid as assembly aceptor vector, and transform the production reaction in E. coli competent cells, you have to plate them in culture media supplemented with kanamycin at 50 μg/mL, X-Gal at 40 μg/mL, and IPTG 0.2 μmol/mL. This last one is necessary to have expresion of the LacZ negative selection marker.
Then you have to select some white colonies (e.g. 3 colonies) and perform a propper assembly verification method like colony PCR or sequencing. If everthing is correct, you can transform the obtained plasmid in your expression strain and carry controled overexpression by induction with IPTG.
I hope it is clear